Human systems biology in real-world settings
Systems biology," which considers life as systems and understands target systems from the characteristics and relationships of their components, has been developed. However, in order to understand higher-order phenomena such as the behavior of mammals, especially humans, at the individual level, it is necessary to take into account the complexity of social structures and the influence of "environmental factors," and this methodology has not yet been fully established. In this project, we will develop human systems biology using sleep-wake rhythms as a model system. We would like to understand the "biological timing" information of human sleep and wakefulness from the molecular level to the level of individuals living in society. In this project, we aim to elucidate the molecular mechanisms of sleep and wakefulness in mammals, including humans, using our human sleep assay and next-generation mouse genetics, and to clarify the causal relationship between gene polymorphisms and phenotypes. Furthermore, we are focusing on the regulation of protein phosphorylation to understand and control sleep-wake rhythms at the molecular level. The "sleep phosphorylation hypothesis" that we propose is the key to clarifying how humans process information about fatigue and sleepiness. In addition, sleep research is expected to provide an important foundation for a better understanding of mental disorders and the development of new treatments.
Ueda Biological Timing PJ
Human systems biology in real-world settings
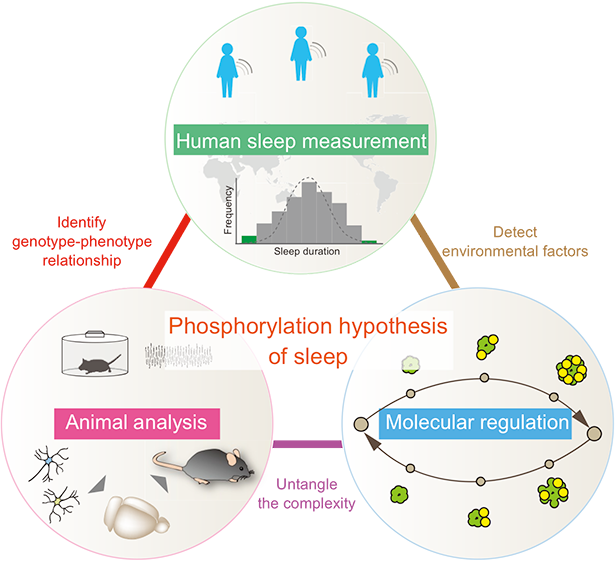
Human Sleep GroupFrom population to correlation
Based on simple and accurate human sleep measurement, the research group aims to draw the landscape of human sleep and extracts rare mutations that cause significant changes in sleep phenotype from a large population.
The research group has successfully developed an algorithm to accurately determine sleep and wakefulness using a wristwatch-type wearable device. Based on this technology, we will proceed with large-scale sleep measurement and challenge the definition of "healthy sleep", which has not been achieved so far.
Animal Analysis GroupFrom correlation to causality
In order to clarify the causal relationship between genetic and environmental factors and sleep time information processing, this group will produce gene knockout, knockin, and overexpressing mice and analyze their sleep phenotypes in parallel. By introducing three guide RNAs and CRISPR/Cas9 into fertilized mouse eggs, the research group has developed a method for obtaining mice in which the genes of both alleles have been knocked out in almost whole body cells in one generation without mating. In addition, we have established an ES mouse method in which almost whole body cells are derived from genetically modified ES cells, and have succeeded in parallel production of gene knock-in mice. The sleep phenotype of genetically modified mice is analyzed using the SSS method, which is a method for determining sleep / wakefulness based on respiratory waveforms.
Molecular Control GroupFrom causality to control
We will elucidate the qualitative control mechanism of sleep control proteins and perform strategies for perturbing them at the single residue level. Changes in the activity of individual gene products (molecules such as proteins) lead to changes in phenotype at the individual level under various influences including cell level, organ level, and even social factors. Therefore, it may be considered that there is an incomprehensible gap between molecular activity and individual phenotype. However, there are some examples, such as the circadian clock, that can accurately understand how changes in molecular activity affect individual phenotypes if the individual phenotype is properly evaluated. What about the sleep-wake rhythms that govern our daily behavior? The molecular control group aims to understand and manipulate the sleep phenotype of mammals, including humans, directly from specific molecule activity changes through the perturbation based on the understanding of protein activity control mechanisms centered on phosphorylation.




