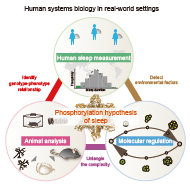
Department of systems pharmacology is conducting research to establish human systems biology and pharmacology using sleep-wake cycle as a model system, which is common to organisms with a central nervous system and one of the main factors that dictate the dynamics of social behavior. With our members from different backgrounds, we would like to realize systems biology at the organism level including human, leading to greater understanding and even control of organismal pathophysiology.
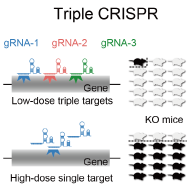
In the conventional method for production of genetically-modified mice, a single line of gene-targeted ES cells is injected into host embryos (typically use blastocysts) to generate chimera mice comprising a mixture of ES- and host-derived cells. In addition, multiple mating procedures are needed to generate the desired genetically modified mouse strain, which typically takes from 9 months to several years. Such multiple rounds of mating procedures are necessary to produce gene knockout / knock-in mice in the conventional method, which was usually a bottleneck to promote organism-level system biology. Therefore, we propose the concept of "next-generation mouse genetics" which creates genetically modified mice without mating and uses it for analysis. We implemented the concept by developing the triple-CRISPR method (Sunagawa et al., Cell Rep. 2016) and the ~100% ES cell-derived mice (ES mice) production method (Ode et al., Mol. Cell 2017; Ukai et al. Nat Protoc. 2017).
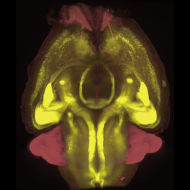
To highlight the regulatory cellular networks in the sleep/wake rhythm, we facilitate identifying sleep/wake regulating cells in the whole-brain in a comprehensive and parallelized manner. CUBIC (Clear, Unobstructed Brain/Body Imaging Cocktails and Computational Analysis) is a great leap forward. It offers a simple tissue clearing protocol by using aminoalcohols and an unprecedented rapid whole-brain and whole-body imaging at single-cell resolution (represented by Susaki et al., Cell, 2014; Tainaka et al., Cell 2014; Tainaka et al., Cell Rep. 2018; Murakami et al., Nat. Neurosci. 2018). The development of the CUBIC method has evolved into the development of microscopes for high-speed imaging (Matsumoto et al., Nat. Protoc. 2019), the extension of technology for immunostaining of three-dimensional tissues (Susaki et al., Nat. Commun. 2020), and the development of a data analysis platform for whole-brain single-cell analysis (Mano et al., Cell Rep. Methods 2021). We have also published results of the application of CUBIC to organs other than the brain through collaborative research.

The comprehensive identification of molecular circuits at the organism level also requires accurate (>90%) phenotype analysis. In neuroscience, sleep/wake behavior is an intriguing phenotype, because sleep disorders (e.g., insomnia or hypersomnia) are sensitive and informative symptoms of almost all psychological disorders. Sleep/wake states have been characterized in humans by electroencephalography (EEG) and electromyography (EMG). Characteristic EEG/EMG patterns during sleep and waking are preserved in mammals and can be measured by electrodes surgically implanted in the brain and muscles. However, such recording requires special surgical skills, and the surgery is highly invasive, requiring a long recovery period (more than 10 days) after implantation before sleep/wake recording. Furthermore, the EEG/EMG data are often manually annotated and classified into sleep/wake phenotypes by visual assessment, which can be time consuming and somewhat subjective. To develop a scalable, non-invasive, fully automated sleep/wake recording method for comprehensive studies, we have developed 1) an automated EEG/EMG analysis software (FASTER) (Sunagawa et al., Genes Cell 2013), 2) a respiration-based, non-invasive, fully automated system (SSS) (Sunagawa et al., Cell Rep. 2016). We then have elaborated our technology platforms for highly precise, large-scale, non-invasive measurements of sleep patterns, derived in mice, to humans. We developed a novel algorithm, ACCEL, which classifies sleep and wake with high sensitivity and specificity by using jerk of arm movement recorded by wristwatch-type accelerometers (Ode et al., iScience 2022).
Sleep amount during a day is under homeostatic control, in which sleep pressure accumulates during awake time and gradually decreases during sleep. Sleep deprivation further promotes the accumulation of sleep pressure, resulting in the longer/deeper sleep in the next cycle. The required sleep duration in a day is mostly determined genetically as evident from the fact that each animal spices shows characteristic different sleep duration. However, detailed molecular mechanisms underlying the control of sleep duration in mammals are still elusive. Using triple-target CRISPR, SSS, and CUBIC techniques together with a computational model that simulates the membrane potential of cortex neuron regulated by a group of ion channels, we have discovered and proposed that Ca2+-dependent neuronal hyperpolarization pathway affects sleep duration in mammals (Sunagawa et al., Cell Reports 2016; Tatsuki et al., Neuron 2016; Yoshida et al. PNAS 2019; Yamada et al., iScience 2022).
We have further been investigating molecules that regulate the transition and consolidation of sleep and awake phases. In this regard, we have reported a phenotype of Chrm1 and Chrm3 double-knockout mice produced by triple-CRISPR technology, which showed a complete loss of REM sleep (Niwa et al., Cell Rep. 2018; Yamada and Ueda, Front. Neurosci. 2020). These results were the first report of essential genes of REM sleep. This study also indicated that REM sleep, defined through EEG-EMG recordings, is not critical for the survival of animals.
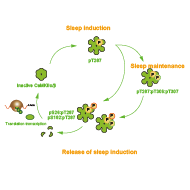
In the process of investigating the relationship between Ca2+-dependent regulation of neuronal activity and sleep, we discovered CaMKIIα/β as the first sleep-inducing protein kinase in mammals (Tatsuki et al., Neuron 2016). Since then, the relationship between protein phosphorylation and sleep regulation has been suggested by multiple genetic and biochemical findings—leading to the proposal of phosphorylation hypothesis of sleep (Ode and Ueda, Front. Psychol. 2021). Phosphorylation/dephosphorylation-mediated protein regulation represents a promising target in sleep–wake regulation because it can occur on variable dynamics and time scale from minutes to hours, and allows for the integration of genetic factors (regulation by amino acid sequence) and environmental factors (posttranslational modification of intracellular signaling pathways).
Do molecular mechanisms at the single amino acid residue level, such as specific protein phosphorylation, control the organism-level phenotype including human sleep-wake cycle? We will ask this question leveraging mouse genetics as a springboard to validate genotype–phenotype relationships.
We have elaborated our technology platforms for highly precise, large-scale, non-invasive measurements of sleep patterns, derived in mice, to humans. We developed a novel algorithm, ACCEL, which classifies sleep and wake with high sensitivity and specificity by using jerk of arm movement recorded by wristwatch-type accelerometers (Ode et al., iScience 2022). Using ACCEL, we estimated the sleep-wake dynamics from the arm acceleration data of 100,000 people registered in the UK biobank, and succeeded in classifying human sleep patterns into 16 types (Katori et al., PNAS 2022). Based on the ACCEL and sleep classification methods, we are working on social implementation of "sleep checkup" that will allow many people in our society to understand their own sleep status and trigger necessary changes in their own sleep behavior.
Humans provide a rich source of information on the roles of genetic and environmental factors in sleep–wake control. We hope that our research results will contribute to the understanding of sleep disorders, circadian rhythm disorders, and associated psychiatric and neurodegenerative diseases, and to the search for therapeutic strategies.